Enriched zones of embedded ribonucleotides are associated with DNA replication and coding sequences in the human mitochondrial genome
Abstract
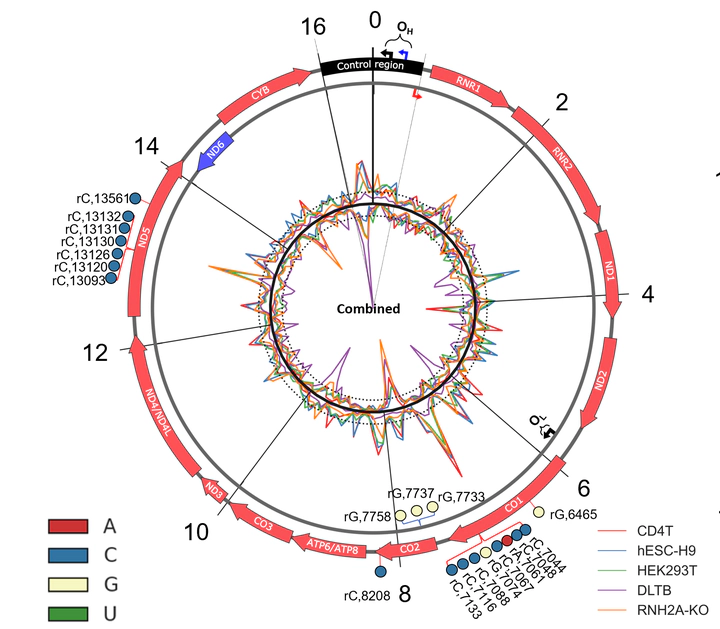
Abundant ribonucleoside triphosphate (rNTP) incorporation in DNA by DNA polymerases in the form of ribonucleoside monophosphates (rNMPs) is a widespread phenomenon in nature, resulting in DNA structural change and genome instability. The rNMP distribution, characteristics, hotspots, and association with DNA metabolic processes in human mitochondrial DNA (hmtDNA) remain mostly unknown. Here, we utilize the ribose-seq technique to capture embedded rNMPs in mtDNA of six different human cell types with wild-type or mutant ribonuclease (RNase) H2 genotype. The rNMPs are preferentially embedded in the DNA of the light strand in most cell types studied, but not in the liver-tissue cells, in which the rNMPs are dominant on the heavy strand of hmtDNA. We uncover common rNMP hotspots and conserved rNMP-enriched zones across the entire hmtDNA, including in the replication-control region, which may result in the suppression of mtDNA replication. We also show that longer coding sequences have a significantly higher rNMP-embedment frequency per nucleotide. While the composition of the embedded rNMPs varies among the different cell types, by studying the genomic context of embedded rNMPs, we detected common rNMP-embedment patterns in hmtDNA. The genomic contexts of rNMPs found in hmtDNA are mainly distinct from those found in yeast mtDNA, highlighting a unique signature of rNTP incorporation by hmtDNA polymerase γ.Competing Interest StatementThe authors have declared no competing interest.