Optimization of rNMP capture techniques
Description
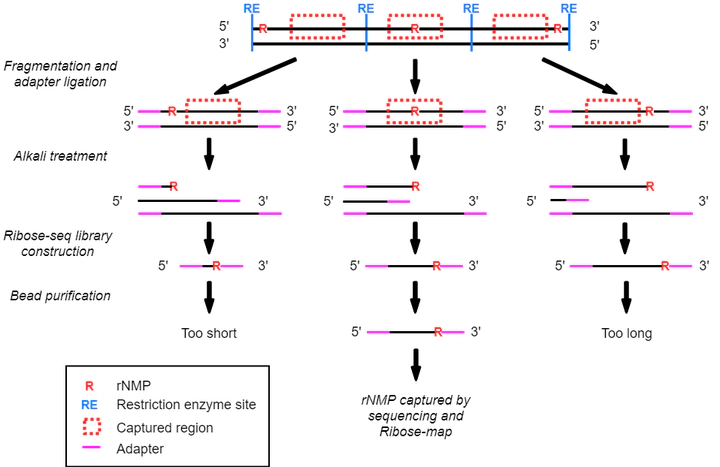
Ribonucleoside triphosphates (rNTPs) are incorporated into DNA by DNA polymerases in the form of ribonucleoside monophosphates (rNMPs), which are the most abundant non-standard nucleotides in genomic DNA in many species. The presence of rNMPs in DNA leads to DNA structural change, genome instability, and affects protein-DNA interactions. The rNMP incorporation is also associated with a bunch of human disease including Aicardi-Goutières Syndrome and various type of cancers. Since the last decade, researchers have developed a series of rNMP mapping techniques. However, compared to the high rNMP incorporation frequency, the efficiency and coverage of rNMP mapping techniques are still lower than research needs. In this study, we aim to improve the both efficiency and the coverage of rNMP mapping techniques by optimizing the current rNMP-mapping technique with in silico simulation and develop novel rNMP mapping technique with the help of modern biological techniques.
Funding sources
- HHMI 55108574 Mechanisms of RNA-mediated genomic stability and instability, Howard Hughes Medical Institute Faculty Scholars Award (Francesca Storici, PI)
- R01 ES026243 Ribose-seq profile and analysis of ribonucleotides in DNA of oxidatively-stressed and cancer cells, _National Institute of Health (NIH) NIEHS (Francesca Storici, PI; Gianluca Tell, subcontract co-Investigator)